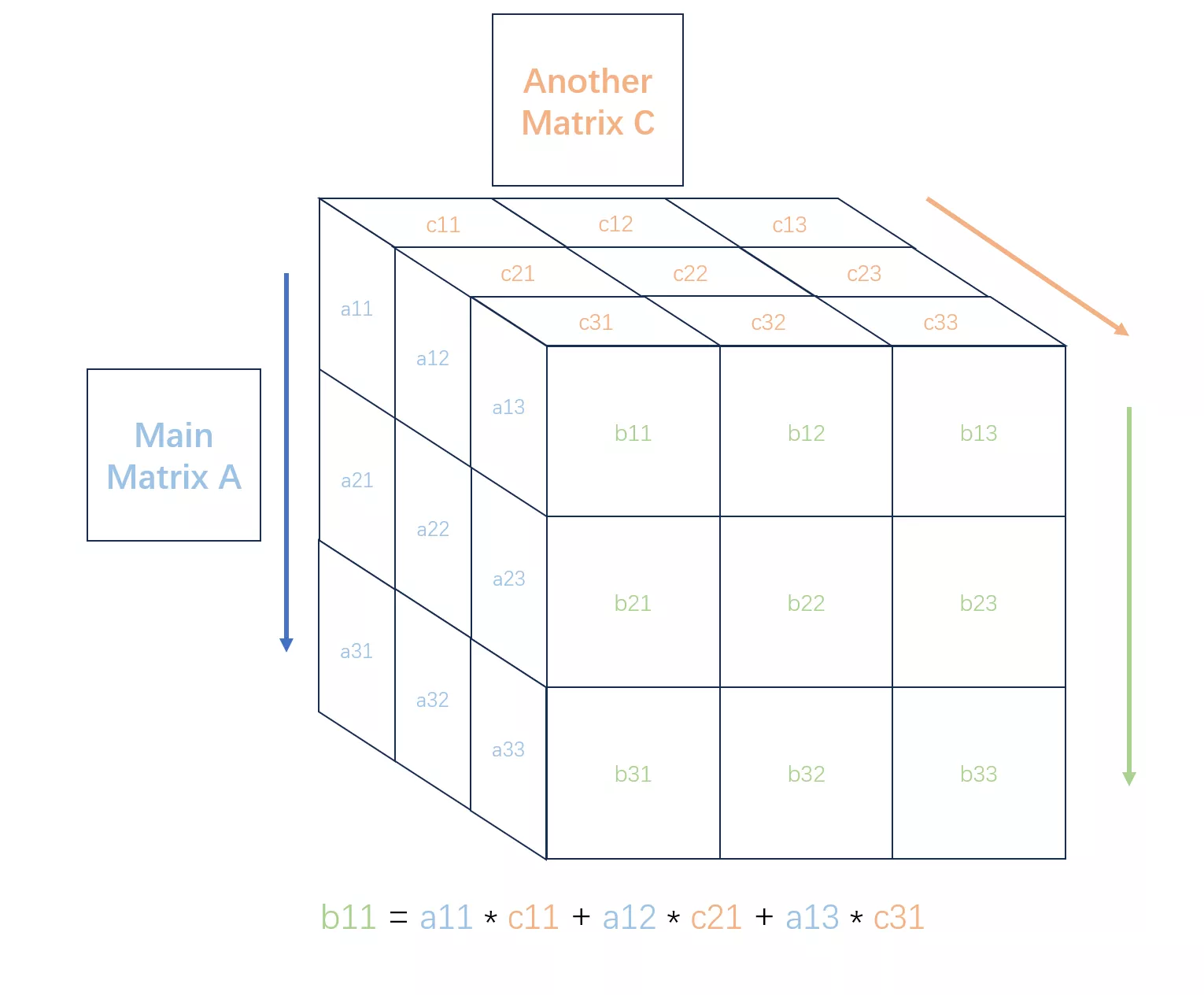
In this article, I will try to replicate the insurance pricing models with freMTPL2 Dataset from: https://www.kaggle.com/code/floser/use-case-claim-frequency-modeling-python/notebook#3.-Model-champions:-Gradient-tree-boosting-machines. I also find some coding errors and will try to fix it.
Coding
Packages
No special environment needed. We use normal packages to replicate the model.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import statsmodels.api as sm
import statsmodels.formula.api as smf
from catboost import Pool, CatBoostRegressor, cv
from lightgbm import LGBMRegressor
from xgboost import XGBRegressor
import time
from sklearn.model_selection import GridSearchCV
# Explainable AI methods
import shap
shap.initjs()We also need to set the seeds and plot theme. Here I simplify some setting and dismiss the device option.
seed = 123
sns.set(rc={'figure.figsize':(12,7)})
sns.set_theme()Helper function
Here we define some functions so that it could be used to verify our results. I have modified from the original article. Now the function has less parameters and could be used easily.
# Modified function PDW: Weighted Poisson Deviance
def PDW(df, prediction):
pred = df[prediction]
obs = df['ClaimNb']
ex = df['Exposure']
deviance = 200 * np.sum(ex * (pred - obs + np.log((obs / pred) ** obs))) / np.sum(ex)
return deviance
# Function PDW2: Print Poisson Deviance learn/test
def PDW2(txt, df_learn, df_test, prediction):
print("%s, Learn/Test: %.2f %% / %.2f %%" % (txt, PDW(df_learn,prediction), PDW(df_test,prediction)))
# Function CF2: Print claim frequency
def CF2(txt, train, test, target_train='ClaimNb', target_test='ClaimNb'):
train_freq = train[target_train].sum()/train['Exposure'].sum()*100
test_freq = test[target_test].sum()/test['Exposure'].sum()*100
print("%s: %.2f %% / %.2f %%" % (txt, train_freq, test_freq))
# Function Benchmark.GLM2: Improvement in Poisson Deviance on test set compared to GLM2-INT-Improvement
def Benchmark_GLM2(txt, df, pred):
# print(PDW(df, pred))
# print(PDW(df, 'claim_freq_with_e'))
# print(PDW(df, 'fitGLM2_with_e'))
index = (PDW(df, pred) - PDW(df, 'claim_freq_with_e'))/ (PDW(df, 'fitGLM2_with_e') - PDW(df, 'claim_freq_with_e')) * 100
II.append(index)
mname.append(txt)
print("GLM2-Improvement-Index (PD test) of %s: %.1f %%" % (txt, index))
def plot_eval(d,x1,x2,t):
"""
This function graphically represents the forecast quality of the fitted models.
Input parameter: Dictionary with model names, PD and Text
"""
df_eval = pd.DataFrame(d)
sns.set_style('darkgrid')
plt.title("GLM2-Improvement-Index of the fitted models: " + t)
sns.barplot(data = df_eval, x = "improvement", y = "modelname", color = 'magenta')
plt.xlim(x1, x2)
plt.show()
def plot_feature_importance(model, data, top_n=20, model_name=''):
"""
This function extracts and plots the most important features (if method feature_importance_ is available)
"""
# Extract the most important features and create a Series for plotting
FI = pd.Series(model.feature_importances_, index=data.columns).nlargest(top_n)
# Plot the feature importances
FI.plot(kind='barh')
plt.gca().invert_yaxis()
text = 'Model {}: Top {} most important features'.format(model_name, top_n)
plt.title(text)
plt.xlabel('Importance')
plt.show()
def range1(df, prediction):
# Sort the DataFrame by the prediction column
pred = prediction
df['sort'] = df[pred]/df['Exposure']
df_sorted = df.sort_values(by='sort').reset_index(drop=True)
# Group the data into deciles
try:
df_sorted['decile'] = pd.qcut(df_sorted['sort'], 10, labels=False) + 1
except ValueError as e:
print(f"Error when grouping with qcut: {e}")
# If qcut fails, use cut for equal-width grouping
df_sorted['decile'] = pd.cut(df_sorted['sort'], bins=10, labels=False) + 1
# Calculate the sum of ClaimNb for the first and last deciles
df_1 = df_sorted[df_sorted['decile'] == 1]
claim_sum_first_decile = df_1['ClaimNb'].sum() / df_1['Exposure'].sum()
df_10 = df_sorted[df_sorted['decile'] == 10]
claim_sum_last_decile = df_10['ClaimNb'].sum() / df_10['Exposure'].sum()
# Output the results
print(f"Sum of ClaimNb for first decile (1st decile): {claim_sum_first_decile:.2%}")
print(f"Sum of ClaimNb for last decile (10th decile): {claim_sum_last_decile:.2%}")
# Optional: View the range between the last and first decile
range_1 = claim_sum_last_decile / claim_sum_first_decile
print(f'Range: {range_1:.2f}')Data preparing
I recommend to use Kaggle command to download all the materials. You could find the command from the reference above.
path = kagglehub.dataset_download("floser/fremtpl2sev")
print("Path to dataset files:", path)After downloading all three csv documents, we will clean them and do some feature engineering works.
# Put all csv documents in data folder
df_freq = pd.read_csv('data/freMTPL2freq.csv')
df_sev = pd.read_csv('data/freMTPL2sev.csv')
df_folds = pd.read_csv('data/freMTPL2freq_folds.csv')
# Format changes
df_freq.IDpol = df_freq.IDpol.astype(int) # policy ID should be integer
df_folds.IDpol = df_folds.IDpol.astype(int) # policy ID should be integer
# Merge freq and folds no. so that we will have the same train/test sets
df = pd.merge(df_freq, df_folds, how='inner', on='IDpol')
print("Number of raw data rows and columns: ", df_freq.shape)
print("Number of data rows and columns: ", df.shape)
print("Check the end of the data set (possibly corrupted data):")
df.tail(6) # view end of dataset
# Merge freq and sev so that we combine the policies and claims in one table
df_sev["ClaimNb"] = 1
sev_agg = df_sev.groupby("IDpol").sum("ClaimNb").drop(columns=['ClaimAmount'])
# left join with claims frequency data (exposures, features and without questionable ClaimNb).
df = pd.merge(df.drop(columns=['ClaimNb']), sev_agg, on="IDpol", how="left").fillna(0)
# data preparations and corrections as used in Schelldorfer and Wüthrich (2019)
df['ClaimNb'] = df['ClaimNb'].clip(upper=4)
df['Exposure'] = df['Exposure'].clip(upper=1)
# count number of missing values in data set
print('Null Count:', df.isna().sum().sum())
# Show summary statistics
df.describe()
df.info()
df['Frequency'] = df['ClaimNb'] / df['Exposure']Now prepare two different datasets. One for GLM and the other for ML/DL. We also prepare a test set to store all results.
# prepare data for GLM modeling
df_GLM = df.copy()
df_GLM["AreaGLM"] = df_GLM.Area.factorize()[0]
df_GLM["VehPowerGLM"] = np.minimum(9, df_GLM.VehPower)
# VehAgeGLM: 3 classes
mapping = {k: 3 if k>=11 else (2 if k>= 1 else 1) for k in range(111)}
df_GLM["VehAgeGLM"] = df_GLM.VehAge.map(mapping)
df_GLM["BonusMalusGLM"] = np.minimum(150, df_GLM.BonusMalus)
df_GLM["DensityGLM"] = np.log(df_GLM.Density)
df_GLM.head()
# Defining learn and test sample based on previously defined folds
train_GLM = df_GLM.loc[df_GLM.fold != 5,].copy() # 80%
test_GLM = df_GLM.loc[df_GLM.fold == 5,].copy() # 20%
CF2("Claim Frequency (Actual) Learn/Test", train_GLM, test_GLM)
# Model INT "predictions"
claim_freq = train_GLM['ClaimNb'].sum() / train_GLM['Exposure'].sum() # claim frequency
train_GLM["claim_freq_with_e"] = claim_freq * train_GLM['Exposure']
test_GLM["claim_freq_with_e"] = claim_freq * test_GLM['Exposure']
df["claim_freq_with_e"] = claim_freq * df['Exposure']
# Print claim frequency actual vs predicted (Intercept model)
CF2("Claim Frequency INT, Test-Sample, Actual/Predicted", test_GLM, test_GLM, target_test='claim_freq_with_e')
# Print Poisson Deviance
PDW2("Poisson Deviance INT", train_GLM, test_GLM, "claim_freq_with_e")
# prepare test set
test_set = df.loc[df_GLM.fold == 5,['IDpol','Exposure','ClaimNb']].copy()
test_set["claim_freq_with_e"] = test_GLM["claim_freq_with_e"]
# Convert categorical variable into dummy/indicator variables.
df_onehot = pd.get_dummies(df, drop_first=False, dtype='int')
df_onehot["claim_freq_with_e"] = claim_freq * df_onehot['Exposure']
print(df_onehot.shape)
df_onehot.head()
# generate sample with encoded data
learn_onehot = df_onehot.loc[df_onehot.fold != 5,].copy() # 80%
test_onehot = df_onehot.loc[df_onehot.fold == 5,].copy() # 20%
# Additionally, let's divide learn into train and validation (e.g. to use it for early stopping)
train_onehot = df_onehot.loc[df_onehot.fold < 4,].copy() # 60%
val_onehot = df_onehot.loc[df_onehot.fold == 4,].copy() # 20%
CF2("Claim Frequency (Actual) Train/Val", train_onehot, val_onehot)
# ML-convention: Generate separate datasets for labels (y) and features (X)
no_feature_list=['IDpol', 'ClaimNb', 'Exposure', 'Frequency', 'fold', 'claim_freq_with_e']
# labels
y_train = train_onehot.ClaimNb
y_val = val_onehot.ClaimNb
y_test = test_onehot.ClaimNb
y_learn = learn_onehot.ClaimNb
# features
X_train = train_onehot.drop(no_feature_list, axis=1)
X_val = val_onehot.drop(no_feature_list, axis=1)
X_test = test_onehot.drop(no_feature_list, axis=1)
X_learn = learn_onehot.drop(no_feature_list, axis=1)
# Get feature names from encoded data
feature_names = X_learn.columns
# Display first observations of the learning sample
X_learn.head(3)Modeling - GLM
We modeled several GLM models.
# Vanila GLM
model = smf.glm("ClaimNb ~ C(VehPowerGLM) + C(VehAgeGLM) + BonusMalusGLM + VehBrand + VehGas + DensityGLM + C(Region) + AreaGLM + DrivAge + np.log(DrivAge) + I(DrivAge**2) + I(DrivAge**3) + I(DrivAge**4)", family=sm.families.Poisson(), offset = np.log(train_GLM.Exposure), data=train_GLM)
# Optimizer: https://www.statsmodels.org/dev/generated/statsmodels.discrete.discrete_model.Poisson.fit.html
d_glm2 = model.fit(method='newton') # same results with 'nm', 'IRLS'
#d_glm2.summary()
# make predictions
train_GLM["fitGLM2_with_e"] = d_glm2.predict(train_GLM) * train_GLM.Exposure
test_GLM["fitGLM2_with_e"] = d_glm2.predict(test_GLM) * test_GLM.Exposure
test_set["fitGLM2_with_e"] = test_GLM["fitGLM2_with_e"]
df["fitGLM2_with_e"] = d_glm2.predict(df_GLM) * df.Exposure
# Print claim frequency actual vs predicted
CF2("Claim Frequency GLM2, Test-Sample, Actual/Predicted", test_GLM, test_GLM, target_test='fitGLM2_with_e')
# Print Poisson Deviance
PDW2("Poisson Deviance GLM2", train_GLM, test_GLM, 'fitGLM2_with_e')
range1(train_GLM,'fitGLM2_with_e')
range1(test_GLM,'fitGLM2_with_e')
model = smf.glm("ClaimNb ~ AreaGLM + C(VehPowerGLM) * C(VehAgeGLM) + C(VehAgeGLM) * VehBrand + VehGas * C(VehAgeGLM) + DensityGLM + C(Region) + BonusMalusGLM * np.log(DrivAge) + np.log(BonusMalusGLM) + I(BonusMalusGLM**2) + I(BonusMalusGLM**3) + I(BonusMalusGLM**4) + DrivAge + I(DrivAge**2) + I(DrivAge**3) + I(DrivAge**4)", family=sm.families.Poisson(), offset = np.log(train_GLM.Exposure), data=train_GLM)
d_glm4 = model.fit(method='newton')
#print(d_glm4.summary())
train_GLM["fitGLM4_with_e"] = d_glm4.predict(train_GLM) * train_GLM.Exposure
test_GLM["fitGLM4_with_e"] = d_glm4.predict(test_GLM) * test_GLM.Exposure
test_set["fitGLM4_with_e"] = test_GLM["fitGLM4_with_e"]
# Print Poisson Deviance
PDW2('Poisson Deviance GLM4', train_GLM, test_GLM, 'fitGLM4_with_e')
# Improvement in Poisson Deviance on test set compared to GLM2-INT-Improvement
Benchmark_GLM2("GLM4", test_set, "fitGLM4_with_e")
range1(train_GLM,'fitGLM4_with_e')
range1(test_GLM,'fitGLM4_with_e')Modeling - GBDT
# GBDT
# generate catboost specific "pool" data based on encoded data (ML-standard. CatBoost could process not encoded data)
learn_pool = Pool(data=X_learn, label=y_learn/learn_onehot.Exposure, weight=learn_onehot.Exposure)
train_pool = Pool(data=X_train, label=y_train/train_onehot.Exposure, weight=train_onehot.Exposure)
val_pool = Pool(data=X_val, label=y_val/val_onehot.Exposure, weight=val_onehot.Exposure)
test_pool = Pool(data=X_test, label=y_test/test_onehot.Exposure, weight=test_onehot.Exposure)
model_cb1 = CatBoostRegressor(objective='Poisson', random_seed=seed)
tic = time.time()
model_cb1.fit(learn_pool, logging_level='Silent')
print("time (sec):" + "%6.0f" % (time.time() - tic))
print(model_cb1.get_all_params())
learn_onehot["fitCB1_with_e"] = model_cb1.predict(learn_pool) * learn_onehot.Exposure
test_onehot["fitCB1_with_e"] = model_cb1.predict(test_pool) * test_onehot.Exposure
test_set["fitCB1_with_e"] = test_onehot["fitCB1_with_e"]
# Print Poisson Deviance
PDW2('Poisson Deviance CB1-unconstraint', learn_onehot, test_onehot, 'fitCB1_with_e')
# Improvement in Poisson Deviance on test set compared to GLM2-INT-Improvement
Benchmark_GLM2("CB1-unconstraint", test_set, 'fitCB1_with_e')
range1(learn_onehot,'fitCB1_with_e')
range1(test_onehot,'fitCB1_with_e')
plot_feature_importance(model_cb1, X_learn, 10, "CB1-unconstraint")
# define SHAP tree explainer
explainer1 = shap.TreeExplainer(model_cb1)
shap_values1 = explainer1.shap_values(X_test)
# summarize the effects of all the features
shap.summary_plot(shap_values1, X_test, max_display=10)
# visualize the first prediction's explanation (on log scale)
shap.force_plot(explainer1.expected_value, shap_values1[0,:], X_test.iloc[0,:], matplotlib=True)
# Create monotonic constraints vector: increasing: +1, decreasing: -1, unconstraint: 0
mtc = np.zeros(len(X_learn.columns))
mtc[3] = 1 # BonusMalus increasing
X_learn.iloc[0:1, 3] # Check position in data frame
model_cb2 = CatBoostRegressor(objective='Poisson', monotone_constraints = mtc, random_seed=seed)
tic = time.time()
model_cb2.fit(learn_pool, logging_level='Silent')
print("time (sec):" + "%6.0f" % (time.time() - tic))
print(model_cb2.get_all_params())
learn_onehot["fitCB2_with_e"] = model_cb2.predict(learn_pool) * learn_onehot.Exposure
test_onehot["fitCB2_with_e"] = model_cb2.predict(test_pool) * test_onehot.Exposure
test_set["fitCB2_with_e"] = test_onehot["fitCB2_with_e"]
# Print Poisson Deviance
PDW2('Poisson Deviance CB2', learn_onehot, test_onehot, 'fitCB2_with_e')
# Improvement in Poisson Deviance on test set compared to GLM2-INT-Improvement
Benchmark_GLM2("CB2", test_set, 'fitCB2_with_e')
range1(learn_onehot,'fitCB2_with_e')
range1(test_onehot,'fitCB2_with_e')
model_cb3 = CatBoostRegressor(objective='Poisson', early_stopping_rounds=20, monotone_constraints = mtc, random_seed=seed)
tic = time.time()
model_cb3.fit(train_pool, eval_set=val_pool, logging_level='Silent')
print("time (sec):" + "%6.0f" % (time.time() - tic))
print("best iteration:", model_cb3.get_best_iteration())
train_onehot["fitCB3_with_e"] = model_cb3.predict(train_pool) * train_onehot.Exposure
test_onehot["fitCB3_with_e"] = model_cb3.predict(test_pool) * test_onehot.Exposure
test_set["fitCB3_with_e"] = test_onehot["fitCB3_with_e"]
# Print Poisson Deviance
PDW2('Poisson Deviance CB3 early stopping', train_onehot, test_onehot, 'fitCB3_with_e')
# Improvement in Poisson Deviance on test set compared to GLM2-INT-Improvement
Benchmark_GLM2("CB3 early stopping", test_set, 'fitCB3_with_e')
range1(train_onehot,'fitCB3_with_e')
range1(test_onehot,'fitCB3_with_e')
# Hyperparameter search range for CatBoost
param_grid_CB ={'learning_rate': [0.07,0.1,0.15], 'depth':[3,4] }
# Define variables for results reports
sel_params_CB = ['param_learning_rate','param_depth','mean_test_score','rank_test_score']
# CatBoostRegressor: Hyper parameter optimization with GridSearchCV
tic = time.time()
CB_rs = GridSearchCV(CatBoostRegressor(iterations=500, objective='Poisson', monotone_constraints = mtc),
param_grid_CB, cv=4, n_jobs=-1)
CB_rs.fit(X=X_learn, y=y_learn/learn_onehot.Exposure, sample_weight=learn_onehot.Exposure, logging_level='Silent')
# Print the runtime as well as the five best hyper parameter constellations
print("time (sec):" + "%6.0f" % (time.time() - tic))
print("Best hyper parameters:",CB_rs.best_params_)
pd.DataFrame(CB_rs.cv_results_)[sel_params_CB].sort_values("rank_test_score").head()
# Create new model on all folds with the best parameters
model_cb4 = CatBoostRegressor(**CB_rs.best_params_, n_estimators=500,
objective="Poisson", thread_count=-1,
monotone_constraints=mtc, random_state=seed)
model_cb4.fit(X=X_learn, y=y_learn/learn_onehot.Exposure, sample_weight=learn_onehot.Exposure, logging_level='Silent')
# make predictions
learn_onehot["fitCB4_with_e"] = model_cb4.predict(X_learn) * learn_onehot.Exposure
test_onehot["fitCB4_with_e"] = model_cb4.predict(X_test) * test_onehot.Exposure
test_set["fitCB4_with_e"] = test_onehot["fitCB4_with_e"]
# Print Poisson Deviance
PDW2('Poisson Deviance CB4', learn_onehot, test_onehot, 'fitCB4_with_e')
# Improvement in Poisson Deviance on test set compared to GLM2-INT-Improvement
Benchmark_GLM2("CB4 tuned", test_set, 'fitCB4_with_e')
range1(learn_onehot,'fitCB4_with_e')
range1(test_onehot,'fitCB4_with_e')
# XGB
# Get feature names from the DataFrame
feature_names = X_learn.columns
# Define monotonic constraints dictionary (xgboost requires a dict.)
mtcd = {feature: 0 for feature in feature_names} # Set all features to 0
mtcd[feature_names[3]] = 1 # Set the fourth feature to +1
X_learn.iloc[0:1, 3] # Check position in data frame
model_xgb = XGBRegressor(device='cpu', objective = 'count:poisson', eval_metric='poisson-nloglik',
tree_method = 'hist', monotone_constraints = mtcd, n_jobs=-1)
tic = time.time()
model_xgb.fit(X_learn, y_learn/learn_onehot.Exposure, sample_weight = learn_onehot.Exposure)
print("time (sec):" + "%6.0f" % (time.time() - tic))
learn_onehot["fitXGB_with_e"] = model_xgb.predict(X_learn) * learn_onehot.Exposure
test_onehot["fitXGB_with_e"] = model_xgb.predict(X_test) * test_onehot.Exposure
test_set["fitXGB_with_e"] = test_onehot["fitXGB_with_e"]
# Print claim frequency actual vs predicted
CF2("Claim Frequency XGB, Test-Sample, Actual/Predicted", test_onehot,test_onehot, target_test='ClaimNb')
# Print Poisson Deviance
PDW2("Poisson Deviance XGB", learn_onehot, test_onehot, 'fitXGB_with_e')
# Improvement in Poisson Deviance on test set compared to GLM2-INT-Improvement
Benchmark_GLM2("XGB", test_set, 'fitXGB_with_e')
range1(learn_onehot,'fitXGB_with_e')
range1(test_onehot,'fitXGB_with_e')
# Hyperparameter search range for lightGBM
param_grid_XGB ={'learning_rate': [0.04,0.05], 'max_depth':[4,5] }
# Variable list for displaying the CV result
sel_params_XGB = ['param_learning_rate','param_max_depth','mean_test_score','rank_test_score']
# XGBRegressor: Hyperparameter tuning with GridSearchCV
tic = time.time()
XGB_rs = GridSearchCV(XGBRegressor(n_estimators=500, tree_method='hist', device='cpu',
objective="count:poisson", eval_metric="poisson-nloglik",
monotone_constraints = mtcd),
param_grid_XGB, cv=4, n_jobs=-1)
XGB_rs.fit(X=X_learn, y=y_learn/learn_onehot.Exposure, sample_weight=learn_onehot.Exposure)
# Output runtime and best parameters (as well as the next best)
print("time (sec):" + "%6.0f" % (time.time() - tic))
print("Best parameters:",XGB_rs.best_params_)
pd.DataFrame(XGB_rs.cv_results_)[sel_params_XGB].sort_values("rank_test_score").head(5)
# XGBRegressor: Hyperparameter tuning with GridSearchCV
tic = time.time()
XGB_rs = GridSearchCV(XGBRegressor(n_estimators=500, tree_method='hist', device='cpu',
objective="count:poisson", eval_metric="poisson-nloglik",
monotone_constraints = mtcd),
param_grid_XGB, cv=4, n_jobs=-1)
XGB_rs.fit(X=X_learn, y=y_learn/learn_onehot.Exposure, sample_weight=learn_onehot.Exposure)
# Output runtime and best parameters (as well as the next best)
print("time (sec):" + "%6.0f" % (time.time() - tic))
print("Best parameters:",XGB_rs.best_params_)
pd.DataFrame(XGB_rs.cv_results_)[sel_params_XGB].sort_values("rank_test_score").head(5)
# Create new model on all folds with the best parameters
model_xgb2 = XGBRegressor(**XGB_rs.best_params_, n_estimators=500, tree_method='hist', device='cpu',
objective="count:poisson", eval_metric="poisson-nloglik",
monotone_constraints=mtcd, n_jobs=-1, random_state=seed)
model_xgb2.fit(X=X_learn, y=y_learn/learn_onehot.Exposure, sample_weight=learn_onehot.Exposure)
# make predictions
learn_onehot["fitXGB2_with_e"] = model_xgb2.predict(X_learn) * learn_onehot.Exposure
test_onehot["fitXGB2_with_e"] = model_xgb2.predict(X_test) * test_onehot.Exposure
test_set["fitXGB2_with_e"] = test_onehot["fitXGB2_with_e"]
# Print Poisson Deviance
PDW2('Poisson Deviance XGB2', learn_onehot, test_onehot, 'fitXGB2_with_e')
# Improvement in Poisson Deviance on test set compared to GLM2-INT-Improvement
Benchmark_GLM2("XGB2 tuned", test_set, 'fitXGB2_with_e')
range1(learn_onehot,'fitXGB2_with_e')
range1(test_onehot,'fitXGB2_with_e')
# LGBM
model_lgbm = LGBMRegressor(device='cpu', objective="poisson", metric="poisson",
monotone_constraints = mtc, n_jobs=-1, verbose=-1)
model_lgbm.fit(X=X_learn, y=y_learn/learn_onehot.Exposure, sample_weight=learn_onehot.Exposure)
# make predictions
learn_onehot["fitLGBM_with_e"] = model_lgbm.predict(X_learn) * learn_onehot.Exposure
test_onehot["fitLGBM_with_e"] = model_lgbm.predict(X_test) * test_onehot.Exposure
test_set["fitLGBM_with_e"] = test_onehot["fitLGBM_with_e"]
# Print Poisson Deviance
PDW2('Poisson Deviance LGBM', learn_onehot, test_onehot, 'fitLGBM_with_e')
# Improvement in Poisson Deviance on test set compared to GLM2-INT-Improvement
Benchmark_GLM2("LGBM", test_set, 'fitLGBM_with_e')
range1(learn_onehot,'fitLGBM_with_e')
range1(test_onehot,'fitLGBM_with_e')
Math Thinkings
Here are some points related to modeling.
Exposure As a Weight
Below are two ways to calculate frequency with exposures and the claim numbers. However, formula 2 is wrong and should not be used.
- $\frac{\sum_{i=1}^N C_i}{\sum_{i=1}^N E_i}$, where $C_i$ represents Claim numbers of policy i and $E_i$ represents Exposure of policy i. N represents the total number of policies.
- $\frac{\sum_{i=1}^N \frac{C_i}{E_i}}{N}$ (Wrong!)
Think of one policy with E=0.001 and C=1. If using formula 2, The frequency will be larger than 1. In fact, formula 2 uses the wrong weights (1 in every policy). We could change total number N to $\frac{E_i}{\sum E_i}$, putting it in the numerator as a weight, and this will have the equivalent form with formula 1.
Coding Errors
Now we talk about the usual problems in Python. In Python, the ^ and ** operators serve fundamentally different purposes. If you use ^ instead of **, sometimes it will not raise any error but the result will be totally different.
** Operator: Exponentiation
Purpose
- The
**operator is used for arithmetic exponentiation, raising a number to the power of another.
Usage
base = 2
exponent = 3
result = base ** exponent # 2 raised to the power of 3
print(result) # Output: 8Key Points
- Numeric Operations: Primarily operates on numerical types (
int,float,complex). - Precedence: Has higher precedence than most other operators (e.g., multiplication, division).
- Right-Associative: In expressions with multiple
**operators, evaluation occurs from right to left.
^ Operator: Bitwise XOR
Purpose
- The
^operator performs a bitwise exclusive OR (XOR) operation between two integers. It compares each bit of two numbers and returns a new number whose bits are set to1where the bits of the operands differ.
Usage
a = 5 # Binary: 0101
b = 3 # Binary: 0011
result = a ^ b # Binary: 0110, which is 6 in decimal
print(result) # Output: 6Key Points
- Bitwise Operation: Works at the binary (bit) level of integer representations.
- Operands: Typically used with integers (
int), but starting from Python 3.10, it can also be used withbytesandbytearrayfor bitwise operations. - Precedence: Lower than arithmetic operators like
+and*, but higher than comparison operators.
I will use some examples to show how ^ will influence the result.
Notice that ^ will also create a unique number (interesting, and I will prove it later):
[{i:i^2} for i in range(10)]Out:
[{0: 2},
{1: 3},
{2: 0},
{3: 1},
{4: 6},
{5: 7},
{6: 4},
{7: 5},
{8: 10},
{9: 11}]
In modeling part, we use code like this to create a similar factor. One of the factor should be useless here because they are somehow equivalent:
model = smf.glm("ClaimNb ~ DrivAge + I(DrivAge^2) ",
family=sm.families.Poisson(),
offset = np.log(learn2.Exposure), data=learn2)Let's see the summary with model.fit(method='newton').summary()
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -2.1147 | 0.023 | -91.130 | 0.000 | -2.160 | -2.069 |
| DrivAge | -0.0129 | 0.003 | -3.703 | 0.000 | -0.020 | -0.006 |
| I(DrivAge ^ 2) | 0.0022 | 0.003 | 0.653 | 0.514 | -0.004 | 0.009 |
The P value of "DrivAge^2" shows that this factor might have been explained by DrivAge.
What if we use **2 instead?
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -1.4122 | 0.062 | -22.866 | 0.000 | -1.533 | -1.291 |
| DrivAge | -0.0424 | 0.003 | -15.999 | 0.000 | -0.048 | -0.037 |
| I(DrivAge ** 2) | 0.0003 | 2.67e-05 | 12.249 | 0.000 | 0.000 | 0.000 |
Wow, it looks nice! The ** operator works!
Let's do some more checks. We could use a float number and see what happens:
smf.glm("ClaimNb ~ I(DensityGLM^2).
PatsyError: Error evaluating factor: TypeError: Cannot perform 'xor' with a dtyped [float64] array and scalar of type [bool]
ClaimNb ~ I(DensityGLM^2)
^^^^^^^^^^^^^^^
Cool! That is actually the wrong operator! And it will really influence our results (GLM2-Improvement Index from 118% to 106% if using ^)